Did:
- Fixed the problem illustrated in the previous State of the Project post, with the child_count and parent_count properties not being picked up properly. Turns out the database we’ve been using does not have all of the attributes that the db had when Phylet was up online, so these properties were missing. I wrote a script to add them to the Neo4j database, using py2neo, the Python <—> Neo4j translator package. The code:
from py2neo import neo4j, cypher
graph_db = neo4j.GraphDatabaseService("http://localhost:7474/db/data/")
#get rid of all duplicate relationships
def unique(lst, type):
if type == 'parents':
parents = []
for rel in lst:
enode = rel.end_node
if enode not in parents:
parents.append(enode)
return parents
else:
children = []
for rel in lst:
snode = rel.start_node
if snode not in children:
children.append(snode)
return children
for id in range(0, 3000):
try:
#get list of relationships
my_node = graph_db.node(int(id))
par = graph_db.match(start_node=my_node, rel_type="STREECHILDOF")
chil = graph_db.match(end_node=my_node, rel_type="STREECHILDOF")
#get lst of unique relationships only
parents = unique(par, 'parents')
children = unique(chil, 'children')
#update properties of node on Neo4j
dic = {}
dic['stree_children'] = len(children)
dic['stree_parents'] = len(parents)
my_node.update_properties(dic)
except:
continue
This can also be accomplished using the Neo4j Cypher language, directly in Neo4j:
START n = node(*)
MATCH nodes<-[:STREECHILDOF]-n
WITH n, count( distinct nodes) as stree_parents
SET n.stree_parents = stree_parents
START n = node(*)
MATCH nodes-[:STREECHILDOF]->n
WITH n, count( distinct nodes) as stree_children
SET n.stree_children = stree_children
- Pushed up changes to bitbucket repository. This included getting Sourcetree set up and getting a basic tutorial written for it (as always, it is slightly more complicated in Windows than in Mac/Linux). I also refined some of the changes made; the changes in index.html may not be necessary. The site works fine now without these changes, though they did cause issues before.
- Made the database we are using (named atol.db) available for download through the bitbucket repository. This is a temporary solution – we are looking to switch out this database for a different one soon.
- Finished draft of tutorial that gets user from beginning to working local version of Phylet, including troubleshooting for the most common problems.
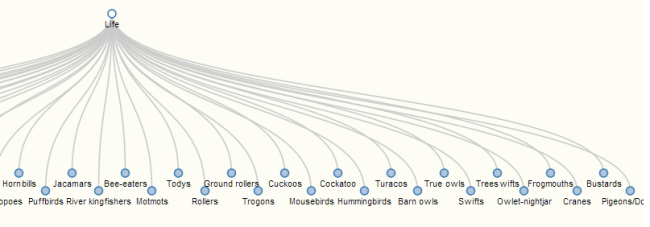
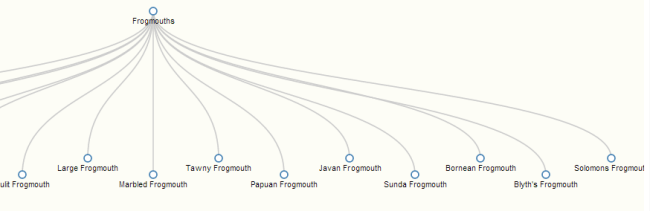
- Began work on a tree visualization (rather than the current force-directed graph). This is a challenge for me. There are many d3 visualizations available online already, complete with code, but they are all written on the assumption that the data is being pulled from a static json file, rather than a database, and that has not been an easy fix for me, as I am still a newbie to both JavaScript and d3. I am currently using this d3 tree visualization as a model. This code builds off of a nested json file that looks a like this:
{
"name": "FLARE",
"children": [
{
"name": "analytics",
"children": [
{
"name": "cluster",
"children": [
{"name": "AgglomerativeCluster", "size": 3938},
{"name": "CommunityStructure", "size": 3812},
{"name": "HierarchicalCluster", "size": 6714},
{"name": "MergeEdge", "size": 743}
]
},
and so on, and so on, with every node including a list of all of its children, and a list of all that child’s children, etc. Assuming the whole Phylet database cannot be loaded all at once because of its size, this necessitates some fudging here. What I’m currently shooting for is to load a node and all of its children at every click. So, starting with the root node “life,” Neo4j would provide all the information for life, and for all of its children. When the user clicks to expand life, Neo4j feeds all of the information for all of the children of those children, so the data is in essence one step ahead of the visualization. So far, I have not been able to get my code to visualize anything at all, though as always it’s impossible to tell if this is because of the visualization itself, or because a connection is broken somewhere. Currently, I am trying to get the working example tree visualization to work inside the Phylet code – my hope is that I will be able to use this strategy to determine when I’ve fixed all the broken connections.
To Do:
- Add the static min version of d3 to the bitbucket repo.
- Continue working on getting alternate visualizations up. My hope is that once I can get one done, doing more will get significantly easier. Current challenges with the tree visualization: even once I get the visualization to work, there may be some problems. Nodes in the Phylet database often have multiple parents, conflicts, etc. that need to be incorporated into the visualization. Nothing in the code I am using now explicitly handles these issues, and I don’t know if it will visualize the multiple-parent issue properly or not. My suspicion is that it will not.
- Help Gabriel flesh out the website as needed.