Documentation on how to set up Neo4j and Phylet is here.
All code will be uploaded to bitbucket here by September 23, 11:59 p.m. EST: https://bitbucket.org/cdphelan/opw-phylet/src/0241cb5e64370faed48ec9cbbc4b77f9c2a3ef01/final_phylet?at=default [sorry – WordPress doesn’t like this as a link for some reason]
Debrief:
The Phylet visualization is now deployable as a visualization tool for researchers. It will probably be most useful to those researchers who are at least somewhat comfortable with fiddling with code. I primarily used the bird database (available as atol.db) in my testing, but it appears to be fairly flexible, as long as the database has a few attributes (given below) Phylet has three primary new features: the tree form, the “breadcrumb” visualization option, and the “backwards” visualization option.
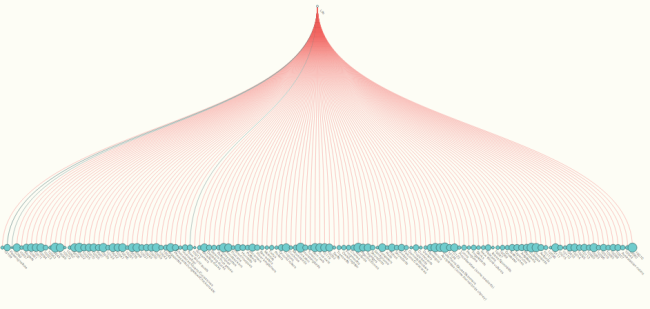
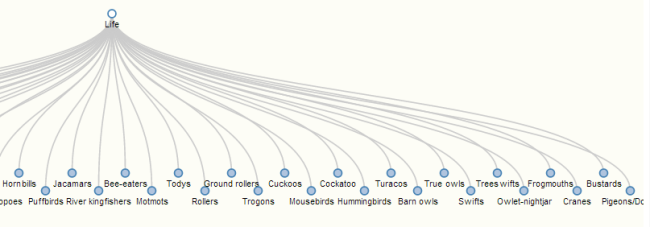
1) Tree form: the visualization is now hierarchical , rather than the free-form force layout from this spring. All nodes start with the life node and branch downwards to a set level; species nodes drop down to a level below all other child nodes, to help with visual organization. Red links indicate conflict, while teal nodes indicate no conflict; the opacity of the link indicates how many sources agree on that connection. Node size correlates with the number of children it has. (It’s fairly easy to rotate this to open from left to right, rather than top to bottom; there are comments in the code to guide this somewhat. )
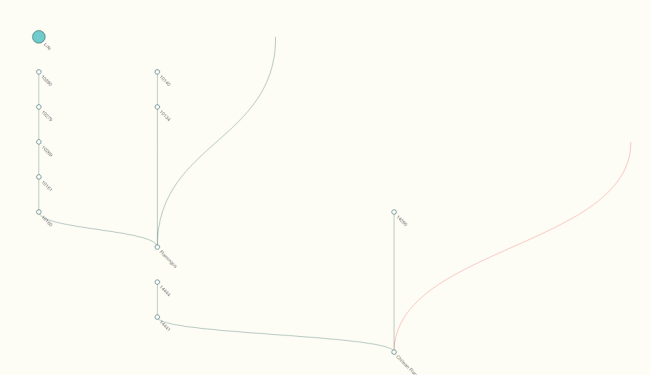
2) Breadcrumb visualization: accessible by right-clicking (or ctrl-click, on a Mac) on a node. In this mode, all expanded nodes collapse except for the clicked node, leaving a single trail from the clicked node back to the root node, Life. One can re-expand any node by left-clicking (with the exception of the bug, see below), or extending the trail by continuing to right-click.
3) Backwards visualization: accessible by shift-clicking a node. This will prompt a separate visualization to pop up that allows a user to explore all of the clicked node’s parents – the clicked node becomes a root node that opens from the bottom up (rather than top down). This functionality is somewhat limited by the multiple-parents problem (see bug report below).
Overall, the result of my summer of coding is what I’d call an “intermediate deliverable”; it is definitely useful, but its functionality is not as broad as it is intended to be. Originally, the intent was to make a visualization that was accessible to both a scientific and general audience, but some work still needs to be done before it can be used by someone without prior understanding of the data structure, without getting them hopelessly confused. The multiple-parent problem causes the nodes and links to act a little strangely, and the number of nodes that are involved in many of these databases makes the full tree fairly overwhelming, visually.
This is a little disappointing for me, of course; I intended to have a complete, deployable visualization ready by the end of the summer. When I set that goal, however, I had no idea what I was getting into – I knew no JavaScript, let alone d3, and my attempts to set up Neo4j on my own were disastrous. It ended up taking me nearly 3 weeks at the beginning of the summer just to get Phylet up and running locally on my machine, which threw off my schedule for the whole summer. It turned out that my difficulty was almost completely attributable to the fact that I was working on a PC – Neo4j appears to have been developed almost exclusively by Mac users, because the three-step installation process on a Mac takes much longer on a PC and requires a lot of fiddling with the code that is not mentioned in the official documentation. The walk-through that I wrote seems to be one of the only such documents online, and I hope it saves at least a few people the process of blind trial-and-error that I had to go through.
Though the results of this summer are different than I planned (which really, is only to be expected), it was unquestionably an incredibly valuable learning experience and for that, I am so grateful. I intend on pursuing a career in data science and particularly data visualization when I complete my masters’ this May, but before this summer I was limited to static visualizations, and was intimidated by the prospect of tackling a project as complex and with as many moving parts as Phylet does. I knew next to nothing about databases, only slightly more about HTML, and was conversationally fluent in Python, but no other programming languages. D3 and JavaScript are incredibly valuable tools to have in my toolbox, and they were challenging enough that I probably would have never really learned them on my own, unless I was forced to.
Sometimes I feel a little guilty – I got so much out of this experience that I feel like it’s not a fair trade; I could never give back as much as I got. Regardless, it was a wonderful experience and I’m glad I had the chance. Thank you to NESCent, Gnome, and particularly Stephen and Gabe, who were wonderful and supportive mentors.
Technical details:
I kept the visualization within the framework of the original Phylet visualization, but of the sake of simplicity, these are the main places where I made changes:
- CSS: various mods to nodes/links
- index.html: uses a static version of d3.min, rather than the ones online; also erased the highlight toggle option
- service.py: primarily used children() and parents(). In children(), there is a count that artificially limits the number of visible children; when the counter reaches a certain number, the loop breaks. You can comment this out/in to turn it off/on (see the code for details).
- gol.js: primarily this.start() and update() and the functions they call; of these, the most important ones that I added are trail(), toggle(), toggleOff(), and load_node().
I made no changes to the search or recreateAction functionalities; I believe they don’t work, perhaps never worked, but making them internet-proof is beyond my expertise. I don’t really know how people use search bars to break sites, for instance.
Requirements for Neo4j format:
The service expects several attributes in a Neo4j node:
- common_name (could easily be replaced with just name)
- stree_children
- stree_parents
The scripts for adding all of these, if they are not already in the database, are available on my bitbucket. My scripts are written in Python, but Neo4j’s cypher language can also be used to do stree_children and stree_parents.
Known bugs:
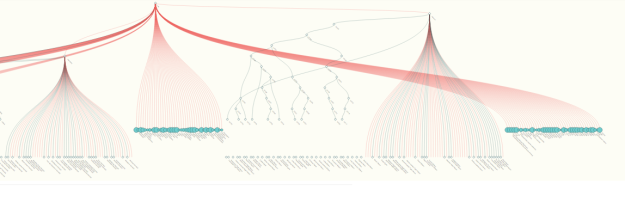
- Multiple parents: The d3 library is not built to handle hierarchical graphs – graphs that are almost trees but have instances where a single node has multiple parents. As far as I can tell, this challenge has not yet been solved by anyone (at least, not anyone who has posted the solution online), and the kind of d3 hacking involved is frankly beyond my JavaScript and d3 capabilities. All discussion centers around making the d3 force layout (which can handle multiple parents) mimic the tree layout (which cannot). I tried this method, but the result is simply not as good as the tree layout, and the physics of the force layout add a lot of bugs into how the force-tree moves.
- Losing children in breadcrumb mode: This happens fairly regularly but I’ve not been able to isolate what causes it to happen. When in “breadcrumb” mode, a user should be able to re-expand nodes by clicking on them, but sometimes this doesn’t work – d3 loses the stored children nodes at some point.
- Window resizing: Minor, but the Phylet website is responsive and dynamically resizes the svg element with the window size; however, the d3 visualization does not resize, which causes the site to look very messy if it’s not exactly the right size. I didn’t attempt to fix this, as I think Gabe has been working on the website look & feel, and didn’t want to fix a problem that may no longer be relevant.
- Space concerns: Not as much of a problem for the research functionality, so it was a low priority and ended up never getting fixed, but the visualization would be fairly unwieldy for a general audience. The visualization requires a lot of space to open fully – there are just too many nodes in a lot of these databases – but I couldn’t find a solution that made Phylet less overwhelming, visually, without impeding its usability for a researcher. I coded, but did not implement, two potential solutions: artificially limiting the number of children a node can have, and creating an additional “load more” node that, upon being clicked, would load n number of nodes more.
- Speed concerns: Opening a node with a lot of children takes about 5-10 seconds, which is a pretty big lag if you don’t know what’s going on. I don’t know how fixable this is.